Frontier Approaches to Whole Brain Emulation (WBE)
Table of Contents
Introduction
Is it possible to upload the brain of an organism onto a computer? This question is the basis of Whole Brain Emulation (WBE), a field which is attempting to do just that.
In pursuit of this, researchers start with a simple type of roundworm known as Caenorhabditis elegans (C. Elegans). This worm is chosen because it’s entire nervous system consisting of 302 neurons has been fully mapped by researchers, including the synaptic connections between the neurons. So in theory, this should allow us to just replicate the worm in-silico and then be done with it?
Well turns out, it’s not that simple. The mapping of the neurons and their connections is known as the connectome. This is akin to knowing the road layout of a city, that doesn’t inform you of the moment to moment traffic conditions, vehicles, pedestrian activity, etc. at any one time.
Just some of the complexities are listed below:
- Synaptic details: The strength of each connection isn’t captured by the basic connectome. Also, each synapse can either be excitatory (encouraging the next neuron to fire), or inhibitory (discouraging the next neuron to fire).
- Dynamic Activity: The strength of connections change over time through processes such as synaptic plasticity, which is fundamental to learning and memory.
- Neuromodulation: The entire nervous system is bathed in a chemical soup of neuromodulators. These substances can change the “mood” of the neural circuits, altering the worm’s behaviour—for example, switching it from a food-seeking state to a resting state—without changing the underlying wiring.
- Embodiment: A brain doesn’t exist in a vacuum. It controls a body with muscles and receives feedback from sensors. To truly emulate the worm, we also need to simulate its physical body and its interaction with the environment.
This means that accurate emulations of an organism and its brain need to account for as many of these properties as possible.
Over time there have been many different projects relating if not directly to whole brain emulation, then to its accompanying fields such as connectomics and fundamental neuroscience research:
OpenWorm Project: An open-source, collaborative project aiming to create the first “digital lifeform” by simulating the C. elegans worm at a cellular level, including its 302 neurons and 95 muscle cells.
Blue Brain Project (BBP): A Swiss-based project (at EPFL) that uses a bottom-up approach to create digital reconstructions and simulations of rodent, and eventually human, brain tissue. It is famous for its detailed simulation of a single rat neocortical column.
Human Brain Project (HBP): A massive, decade-long European research project intended to create a comprehensive digital simulation of the human brain. Its legacy and infrastructure (e.g., simulation tools, atlases) are now continued through the EBRAINS platform.
Virtual FlyBrain: A project focused on the Drosophila (fruit fly), which aims to build a comprehensive, integrated “virtual brain” that combines its connectome (wiring diagram) with gene expression data and other biological information.
IARPA MICrONS: A US government research program that focused on reverse-engineering and mapping a cubic millimeter of a mouse’s visual cortex, a foundational step in gathering the “wiring diagram” data needed for WBE.
However this post will focus on our work reproducing and validating BAAI Worm.
BAAIWorm
BAAIWorm is an in-silico simulation of C. Elegans worms based on an integrative data-driven model consisting of two sub-modules: the brain model and the body-environment model. The goal of the project is re-create C. Elegans chemotaxi behaviour, i.e. the worm moving towards a food source along a gradient.
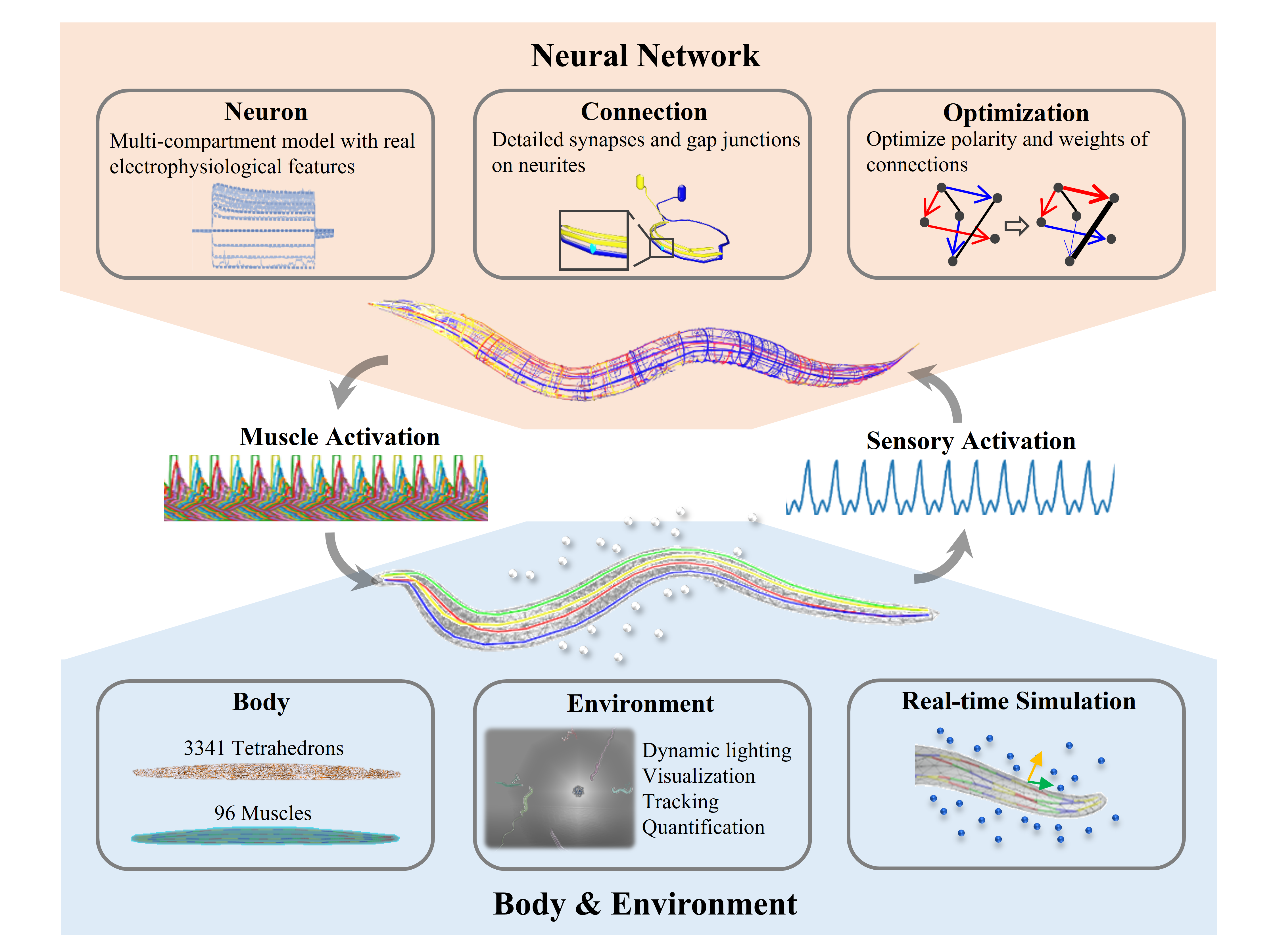
Figure 1: Diagram of BAAIWorm's two component approach to simulating C. Elegans in-silico.
This gives us an interesting simulation of C. Elegans moving in 3-dimensions in a fluid-like space. We have taken the liberty of reproducing the simulation environment from the BAAIWorm paper and instead implementing it as a far more accessible in-browser simulation, check it out below!
Figure 2: Interactive 3D visualization of C. Elegans open-loop simulation. The heatmap shows muscle activation patterns along the body.
The simulation above is animating the exact data readout from the BAAIWorm simulator for it’s open loop demo data from here.
However, the authors of the paper further claim that…
Through the closed-loop interaction between the two submodels, BAAIWorm reproduced the realistic zigzag movement toward attractors observed in C. elegans.
If this is the case, then this represents a useful case of a biorealistic body-environment simulation governed by an equally realistic and plausible biophysical model of C. Elegans chemotaxi behaviour, and the underlying circuits which serve that behaviour.
Thus, this started the journey to reproduce and validate that this is indeed what the project achieved.
Anatomy of Reproduction in Computational Neuroscience
For those of you who are curious about how long it takes to reproduce others findings, here is the timeline for this reproduction and validation effort:
| Epoch | Date Range | Key Activities & Milestones |
|---|---|---|
| 1. Initial Research & Onboarding | July 14 – July 21, 2025 | The project began with a deep dive into the state-of-the-art in Whole Brain Emulation and a thorough review of the BAAIWorm paper. Initial team discussions helped scope the goals for reproduction and validation. |
| 2. Environment Setup & Reproduction | July 24 – Aug 12, 2025 | This phase focused on the significant technical challenge of creating a functional environment. Initial attempts with Docker revealed complex dependencies, leading to a pivot towards a Virtual Machine (VM) that precisely mirrored the original setup. After extensive debugging of core modules (neuron, visualization tools), a stable baseline simulation was finally achieved. |
| 3. Codebase Deep Dive | Aug 13 – Aug 18, 2025 | With a running simulation, the focus shifted to understanding the implementation. This involved a systematic analysis of the codebase, particularly the eworm module, to map out how the brain, body, and environment models were integrated. |
| 4. Porting & Accessibility | Aug 19 – Aug 20, 2025 | An effort was made to increase the model’s accessibility by attempting to port it to macOS and remove its dependency on specific Nvidia hardware. This involved refactoring code, recompiling components, and optimizing performance bottlenecks. |
| 5. Behavioral Validation & Analysis | Aug 21 – Aug 24, 2025 | The core scientific validation began. The primary goal was to replicate the paper’s central claim of chemotaxis (movement towards a food source). Quantitative analysis of the worm’s distance to the food source revealed a consistent divergence from the expected behavior. Debugging focused on the sensory-motor loop, specifically the gradient calculation that guides the worm. |
| 6. Collaborative Review & Findings | August 25, 2025 | The investigation culminated in a productive meeting with one of the paper’s original authors. The findings from the validation analysis were shared and discussed, leading to a deeper understanding of the model’s behavior under the tested conditions and clarifying the path forward. |
The most time consuming aspect of the reproduction was setting up the complex simulation, de-coupling the essential parts from the niceties, and fundamentally understanding the purpose of each component of the entire system w.r.t the published paper.
1. Initial Research and Onboarding
First things first, why did we decide to deep-dive into BAAIWorm in the first place? Based on the papers purported claims, it appeared to be the most biophysically faithful attempt at whole brain emulation out of all of the published attempts based on our initial literature review.
This initial stage involved analysing the research papers claims, with the central one being the quoted chemotaxi behaviour earlier. The team then agreed that we would commit to reproducing the claimed results for the foreesable future.
2. Environment Setup & Reproduction
Setting up and getting the BAAIWorm repo to work was nothing short of painful. This is common with complex, multi-component neuroscience code releases, especially those which weren’t originally intended to be open-sourced.
Resources
Papers
Code
Videos
Exact open-loop simulation in visualisation